Identify Primary Objects identifies biological components of interest in grayscale images containing bright objects on a dark background.
What is a primary object?
In CellProfiler, we use the term
object as a generic term to refer to an identifed feature in an image, usually a cellular subcompartment of some kind (for example, nuclei, cells, colonies, worms). We define an object as
primary when it can be found in an image without needing the assistance of another cellular feature as a reference. For example:
- The nuclei of cells are usually more easily identifiable due to their more uniform morphology, high contrast relative to the background when stained, and good separation between adjacent nuclei. These qualities typically make them appropriate candidates for primary object identification.
- In contrast, cells often have irregular intensity patterns and are lower-contrast with more diffuse staining, making them more challenging to identify than nuclei. In addition, cells often touch their neighbors making it harder to delineate the cell borders. For these reasons, cell bodies are better suited for secondary object identification, since they are best identified by using a previously-identified primary object (i.e, the nuclei) as a reference. See the IdentifySecondaryObjects module for details on how to do this.
What do I need as input?
To use this module, you will need to make sure that your input image has the following qualities:
- The image should be grayscale.
- The foreground (i.e, regions of interest) are lighter than the background.
If this is not the case, other modules can be used to pre-process the images to ensure they are in the proper form:
- If the objects in your images are dark on a light background, you should invert the images using the Invert operation in the ImageMath module.
- If you are working with color images, they must first be converted to grayscale using the ColorToGray module.
If you have images in which the foreground and background cannot be distinguished by intensity alone (e.g, brightfield or DIC images), you can use the ilastik package bundled with CellProfiler to perform pixel-based classification (Windows only). You first train a classifier by identifying areas of images that fall into one of several classes, such as cell body, nucleus, background, etc. Then, the ClassifyPixels module takes the classifier and applies it to each image to identify areas that correspond to the trained classes. The result of ClassifyPixels is an image in which the region that falls into the class of interest is light on a dark background. Since this new image satisfies the constraints above, it can be used as input in IdentifyPrimaryObjects. See the ClassifyPixels module for more information.
What do the settings mean?
See below for help on the individual settings. The following icons are used to call attention to key items:
 Our recommendation or example use case for which a particular setting is best used.
Our recommendation or example use case for which a particular setting is best used. Indicates a condition under which a particular setting may not work well.
Indicates a condition under which a particular setting may not work well. Technical note. Provides more detailed information on the setting.
Technical note. Provides more detailed information on the setting.
What do I get as output?
A set of primary objects are produced by this module, which can be used in downstream modules for measurement purposes or other operations. See the section
"Available measurements" below for the measurements that are produced by this module.
Once the module has finished processing, the module display window will show the following panels:
- Upper left: The raw, original image.
- Upper right: The identified objects shown as a color image where connected pixels that belong to the same object are assigned the same color (label image). It is important to note that assigned colors are arbitrary; they are used simply to help you distingush the various objects.
- Lower left: The raw image overlaid with the colored outlines of the identified objects. Each object is assigned one of three (default) colors:
- Green: Acceptable; passed all criteria
- Magenta: Discarded based on size
- Yellow: Discarded due to touching the border
If you need to change the color defaults, you can make adjustments in File > Preferences. - Lower right: A table showing some of the settings selected by the user, as well as those calculated by the module in order to produce the objects shown.
Available measurements
Image measurements: - Count: The number of primary objects identified.
- OriginalThreshold: The global threshold for the image.
- FinalThreshold: For the global threshold methods, this value is the same as OriginalThreshold. For the adaptive or per-object methods, this value is the mean of the local thresholds.
- WeightedVariance: The sum of the log-transformed variances of the foreground and background pixels, weighted by the number of pixels in each distribution.
- SumOfEntropies: The sum of entropies computed from the foreground and background distributions.
Object measurements:
- Location_X, Location_Y: The pixel (X,Y) coordinates of the primary object centroids. The centroid is calculated as the center of mass of the binary representation of the object.
Technical notes
CellProfiler contains a modular three-step strategy to identify objects even if they touch each other. It is based on previously published algorithms (Malpica et al., 1997; Meyer and Beucher, 1990; Ortiz de Solorzano et al., 1999; Wahlby, 2003; Wahlby et al., 2004). Choosing different options for each of these three steps allows CellProfiler to flexibly analyze a variety of different types of objects. The module has many options, which vary in terms of speed and sophistication. More detail can be found in the Settings section below. Here are the three steps, using an example where nuclei are the primary objects:
- CellProfiler determines whether a foreground region is an individual nucleus or two or more clumped nuclei.
- The edges of nuclei are identified, using thresholding if the object is a single, isolated nucleus, and using more advanced options if the object is actually two or more nuclei that touch each other.
- Some identified objects are discarded or merged together if they fail to meet certain your specified criteria. For example, partial objects at the border of the image can be discarded, and small objects can be discarded or merged with nearby larger ones. A separate module, FilterObjects, can further refine the identified nuclei, if desired, by excluding objects that are a particular size, shape, intensity, or texture.
References
- Malpica N, de Solorzano CO, Vaquero JJ, Santos, A, Vallcorba I, Garcia-Sagredo JM, del Pozo F (1997) "Applying watershed algorithms to the segmentation of clustered nuclei." Cytometry 28, 289-297. (link)
- Meyer F, Beucher S (1990) "Morphological segmentation." J Visual Communication and Image Representation 1, 21-46. (link)
- Ortiz de Solorzano C, Rodriguez EG, Jones A, Pinkel D, Gray JW, Sudar D, Lockett SJ. (1999) "Segmentation of confocal microscope images of cell nuclei in thick tissue sections." Journal of Microscopy-Oxford 193, 212-226. (link)
- Wählby C (2003) Algorithms for applied digital image cytometry, Ph.D., Uppsala University, Uppsala.
- Wählby C, Sintorn IM, Erlandsson F, Borgefors G, Bengtsson E. (2004) "Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections." J Microsc 215, 67-76. (link)
See also IdentifySecondaryObjects, IdentifyTertiaryObjects, IdentifyObjectsManually and ClassifyPixels
Settings:
Select the input image
Select the image that you want to use to identify objects.
Name the primary objects to be identified
Enter the name that you want to call the objects identified by this module.
Typical diameter of objects, in pixel units (Min,Max)
This setting allows the user to make a distinction on the basis of size, which can
be used in conjunction with the
Discard objects outside the diameter range? setting
below to remove objects that fail this criteria.
 The units used here are pixels so that it is easy to zoom in on objects and determine
typical diameters. To measure distances in an open image, use the "Measure
length" tool under Tools in the display window menu bar. If you click on an image
and drag, a line will appear between the two endpoints, and the distance between them shown at the right-most
portion of the bottom panel.
The units used here are pixels so that it is easy to zoom in on objects and determine
typical diameters. To measure distances in an open image, use the "Measure
length" tool under Tools in the display window menu bar. If you click on an image
and drag, a line will appear between the two endpoints, and the distance between them shown at the right-most
portion of the bottom panel.
A few important notes:
- Several other settings make use of the minimum object size entered here,
whether the Discard objects outside the diameter range? setting is used or not:
- Automatically calculate size of smoothing filter for declumping?
- Automatically calculate minimum allowed distance between local maxima?
- Automatically calculate the size of objects for the Laplacian of Gaussian filter? (shown only if Laplacian of
Gaussian is selected as the declumping method)
- For non-round objects, the diameter here is actually the "equivalent diameter", i.e.,
the diameter of a circle with the same area as the object.
Discard objects outside the diameter range?
Select
Yes to discard objects outside the range you specified in the
Typical diameter of objects, in pixel units (Min,Max) setting. Select
No to ignore this
criterion.
Objects discarded
based on size are outlined in magenta in the module's display. See also the
FilterObjects module to further discard objects based on some
other measurement.
 Select Yes allows you to exclude small objects (e.g., dust, noise,
and debris) or large objects (e.g., large clumps) if desired.
Select Yes allows you to exclude small objects (e.g., dust, noise,
and debris) or large objects (e.g., large clumps) if desired.
Try to merge too small objects with nearby larger objects?
Select
Yes to cause objects that are
smaller than the specified minimum diameter to be merged, if possible, with
other surrounding objects.
This is helpful in cases when an object was
incorrectly split into two objects, one of which is actually just a tiny
piece of the larger object. However, this could be problematic if the other
settings in the module are set poorly, producing many tiny objects; the module
will take a very long time trying to merge the tiny objects back together again; you may
not notice that this is the case, since it may successfully piece together the
objects again. It is therefore a good idea to run the
module first without merging objects to make sure the settings are
reasonably effective.
Discard objects touching the border of the image?
Select
Yes to discard objects that touch the border of the image.
Select
No to ignore this criterion.
 Removing objects that touch the image border is useful when you do
not want to make downstream measurements of objects that are not fully within the
field of view. For example, morphological measurements obtained from
a portion of an object would not be accurate.
Removing objects that touch the image border is useful when you do
not want to make downstream measurements of objects that are not fully within the
field of view. For example, morphological measurements obtained from
a portion of an object would not be accurate.
Objects discarded due to border touching are outlined in yellow in the module's display.
Note that if a per-object thresholding method is used or if the image has been
previously cropped or masked, objects that touch the
border of the cropped or masked region may also discarded.
Threshold strategy
The thresholding strategy determines the type of input that is used
to calculate the threshold. The image thresholds can be based on:
- The pixel intensities of the input image (this is the most common).
- A single value manually provided by the user.
- A single value produced by a prior module measurement.
- A binary image (called a mask) where some of the pixel intensity
values are set to 0, and others are set to 1.
These options allow you to calculate a threshold based on the whole
image or based on image sub-regions such as user-defined masks or
objects supplied by a prior module.
The choices for the threshold strategy are:
- Automatic: Use the default settings for
thresholding. This strategy calculates the threshold using the MCT method
on the whole image (see below for details on this method) and applies the
threshold to the image, smoothed with a Gaussian with sigma of 1.
 This approach is fairly robust, but does not allow you to select the threshold
algorithm and does not allow you to apply additional corrections to the
threshold.
This approach is fairly robust, but does not allow you to select the threshold
algorithm and does not allow you to apply additional corrections to the
threshold.
- Global: Calculate a single threshold value based on
the unmasked pixels of the input image and use that value
to classify pixels above the threshold as foreground and below
as background.
 This strategy is fast and robust, especially if
the background is uniformly illuminated.
This strategy is fast and robust, especially if
the background is uniformly illuminated.
- Adaptive: Partition the input image into tiles
and calculate thresholds for each tile. For each tile, the calculated
threshold is applied only to the pixels within that tile.
 This method is slower but can produce better results for non-uniform backgrounds.
However, for signifcant illumination variation, using the CorrectIllumination
modules is preferable.
This method is slower but can produce better results for non-uniform backgrounds.
However, for signifcant illumination variation, using the CorrectIllumination
modules is preferable.
- Per object: Use objects from a prior module
such as IdentifyPrimaryObjects to define the region of interest
to be thresholded. Calculate a separate threshold for each object and
then apply that threshold to pixels within the object. The pixels outside
the objects are classified as background.
 This method can be useful for identifying sub-cellular particles or
single-molecule probes if the background intensity varies from cell to cell
(e.g., autofluorescence or other mechanisms).
This method can be useful for identifying sub-cellular particles or
single-molecule probes if the background intensity varies from cell to cell
(e.g., autofluorescence or other mechanisms).
- Manual: Enter a single value between zero and
one that applies to all cycles and is independent of the input
image.
 This approach is useful if the input image has a stable or
negligible background, or if the input image is the probability
map output of the ClassifyPixels module (in which case, a value
of 0.5 should be chosen). If the input image is already binary (i.e.,
where the foreground is 1 and the background is 0), a manual value
of 0.5 will identify the objects.
This approach is useful if the input image has a stable or
negligible background, or if the input image is the probability
map output of the ClassifyPixels module (in which case, a value
of 0.5 should be chosen). If the input image is already binary (i.e.,
where the foreground is 1 and the background is 0), a manual value
of 0.5 will identify the objects.
- Binary image: Use a binary image to classify
pixels as foreground or background. Pixel values other than zero
will be foreground and pixel values that are zero will be
background. This method can be used to import a ground-truth segmentation created
by CellProfiler or another program.
 The most typical approach to produce a
binary image is to use the ApplyThreshold module (image as input,
image as output) or the ConvertObjectsToImage module (objects as input,
image as output); both have options to produce a binary image. It can also be
used to create objects from an image mask produced by other CellProfiler
modules, such as Morph. Note that even though no algorithm is actually
used to find the threshold in this case, the final threshold value is reported
as the Otsu threshold calculated for the foreground region.
The most typical approach to produce a
binary image is to use the ApplyThreshold module (image as input,
image as output) or the ConvertObjectsToImage module (objects as input,
image as output); both have options to produce a binary image. It can also be
used to create objects from an image mask produced by other CellProfiler
modules, such as Morph. Note that even though no algorithm is actually
used to find the threshold in this case, the final threshold value is reported
as the Otsu threshold calculated for the foreground region.
- Measurement: Use a prior image measurement as the
threshold. The measurement should have values between zero and one.
This strategy can be used to apply a pre-calculated threshold imported
as per-image metadata via the Metadata module.
 Like manual thresholding, this approach can be useful when you are certain what
the cutoff should be. The difference in this case is that the desired threshold does
vary from image to image in the experiment but can be measured using another module,
such as one of the Measure modules, ApplyThreshold or
an Identify module.
Like manual thresholding, this approach can be useful when you are certain what
the cutoff should be. The difference in this case is that the desired threshold does
vary from image to image in the experiment but can be measured using another module,
such as one of the Measure modules, ApplyThreshold or
an Identify module.
Thresholding method
The intensity threshold affects the decision of whether each pixel
will be considered foreground (region(s) of interest) or background.
A higher threshold value will result in only the brightest regions being identified,
whereas a lower threshold value will include dim regions. You can have the threshold
automatically calculated from a choice of several methods,
or you can enter a number manually between 0 and 1 for the threshold.
Both the automatic and manual options have advantages and disadvantages.
 An automatically-calculated threshold adapts to changes in
lighting/staining conditions between images and is usually more
robust/accurate. In the vast majority of cases, an automatic method
is sufficient to achieve the desired thresholding, once the proper
method is selected.
An automatically-calculated threshold adapts to changes in
lighting/staining conditions between images and is usually more
robust/accurate. In the vast majority of cases, an automatic method
is sufficient to achieve the desired thresholding, once the proper
method is selected.- In contrast, an advantage of a manually-entered number is that it treats every image identically,
so use this option when you have a good sense for what the threshold should be
across all images. To help determine the choice of threshold manually, you
can inspect the pixel intensities in an image of your choice.
To view pixel intensities in an open image, use the
pixel intensity tool which is available in any open display window. When you move
your mouse over the image, the pixel intensities will appear in the bottom bar of the display window..
 The manual method is not robust with regard to slight changes in lighting/staining
conditions between images.
The manual method is not robust with regard to slight changes in lighting/staining
conditions between images. - The automatic methods may ocasionally produce a poor
threshold for unusual or artifactual images. It also takes a small amount of time to
calculate, which can add to processing time for analysis runs on a large
number of images.
The threshold that is used for each image is recorded as a per-image
measurement, so if you are surprised by unusual measurements from
one of your images, you might check whether the automatically calculated
threshold was unusually high or low compared to the other images. See the
FlagImage module if you would like to flag an image based on the threshold
value.
There are a number of methods for finding thresholds automatically:
- Otsu: This approach calculates the threshold separating the
two classes of pixels (foreground and background) by minimizing the variance within the
each class.
 This method is a good initial approach if you do not know much about
the image characteristics of all the images in your experiment,
especially if the percentage of the image covered by foreground varies
substantially from image to image.
This method is a good initial approach if you do not know much about
the image characteristics of all the images in your experiment,
especially if the percentage of the image covered by foreground varies
substantially from image to image.
 Our implementation of Otsu's method allows for assigning the threhsold value based on
splitting the image into either two classes (foreground and background) or three classes
(foreground, mid-level, and background). See the help below for more details.
Our implementation of Otsu's method allows for assigning the threhsold value based on
splitting the image into either two classes (foreground and background) or three classes
(foreground, mid-level, and background). See the help below for more details.
- Mixture of Gaussian (MoG):This function assumes that the
pixels in the image belong to either a background class or a foreground
class, using an initial guess of the fraction of the image that is
covered by foreground.
 If you know that the percentage of each image that is foreground does not
vary much from image to image, the MoG method can be better, especially if the
foreground percentage is not near 50%.
If you know that the percentage of each image that is foreground does not
vary much from image to image, the MoG method can be better, especially if the
foreground percentage is not near 50%.
 This method is our own version of a Mixture of Gaussians
algorithm (O. Friman, unpublished). Essentially, there are two steps:
This method is our own version of a Mixture of Gaussians
algorithm (O. Friman, unpublished). Essentially, there are two steps:
- First, a number of Gaussian distributions are estimated to
match the distribution of pixel intensities in the image. Currently
three Gaussian distributions are fitted, one corresponding to a
background class, one corresponding to a foreground class, and one
distribution for an intermediate class. The distributions are fitted
using the Expectation-Maximization algorithm, a procedure referred
to as Mixture of Gaussians modeling.
- When the three Gaussian distributions have been fitted, a decision
is made whether the intermediate class more closely models the background pixels
or foreground pixels, based on the estimated fraction provided by the user.
- Background: This method simply finds the mode of the
histogram of the image, which is assumed to be the background of the
image, and chooses a threshold at twice that value (which you can
adjust with a Threshold Correction Factor; see below). The calculation
includes those pixels between 2% and 98% of the intensity range.
 This thresholding method is appropriate for images in which most of the image is background.
It can also be helpful if your images vary in overall brightness, but the objects of
interest are consistently N times brighter than the background level of the image.
This thresholding method is appropriate for images in which most of the image is background.
It can also be helpful if your images vary in overall brightness, but the objects of
interest are consistently N times brighter than the background level of the image.
- RobustBackground: Much like the Background: method, this method is
also simple and assumes that the background distribution
approximates a Gaussian by trimming the brightest and dimmest 5% of pixel
intensities. It then calculates the mean and standard deviation of the
remaining pixels and calculates the threshold as the mean + 2 times
the standard deviation.
 This thresholding method can be helpful if the majority
of the image is background, and the results are often comparable or better than the
Background method.
This thresholding method can be helpful if the majority
of the image is background, and the results are often comparable or better than the
Background method.
- RidlerCalvard: This method is simple and its results are
often very similar to Otsu.
RidlerCalvard chooses an initial threshold and then iteratively
calculates the next one by taking the mean of the average intensities of
the background and foreground pixels determined by the first threshold.
The algorithm then repeats this process until the threshold converges to a single value.
 This is an implementation of the method described in Ridler and Calvard, 1978.
According to Sezgin and Sankur 2004, Otsu's
overall quality on testing 40 nondestructive testing images is slightly
better than Ridler's (average error: Otsu, 0.318; Ridler, 0.401).
This is an implementation of the method described in Ridler and Calvard, 1978.
According to Sezgin and Sankur 2004, Otsu's
overall quality on testing 40 nondestructive testing images is slightly
better than Ridler's (average error: Otsu, 0.318; Ridler, 0.401).
- Kapur: This method computes the threshold of an image by
searching for the threshold that maximizes the sum of entropies of the foreground
and background pixel values, when treated as separate distributions.
 This is an implementation of the method described in Kapur et al, 1985.
This is an implementation of the method described in Kapur et al, 1985.
- Maximum correlation thresholding (MCT): This method computes
the maximum correlation between the binary mask created by thresholding and
the thresholded image and is somewhat similar mathematically to Otsu.
 The authors of this method claim superior results when thresholding images
of neurites and other images that have sparse foreground densities.
The authors of this method claim superior results when thresholding images
of neurites and other images that have sparse foreground densities.
 This is an implementation of the method described in Padmanabhan et al, 2010.
This is an implementation of the method described in Padmanabhan et al, 2010.
References
- Sezgin M, Sankur B (2004) "Survey over image thresholding techniques and quantitative
performance evaluation." Journal of Electronic Imaging, 13(1), 146-165.
(link)
- Padmanabhan K, Eddy WF, Crowley JC (2010) "A novel algorithm for
optimal image thresholding of biological data" Journal of
Neuroscience Methods 193, 380-384.
(link)
- Ridler T, Calvard S (1978) "Picture thresholding using an iterative selection method",
IEEE Transactions on Systems, Man and Cybernetics, 8(8), 630-632.
- Kapur JN, Sahoo PK, Wong AKC (1985) "A new method of gray level picture thresholding
using the entropy of the histogram." Computer Vision, Graphics and Image Processing,
29, 273-285.
Select binary image
(Used only if Binary image selected for thresholding method)
Select the binary image to be used for thresholding.
Manual threshold
(Used only if Manual selected for thresholding method)
Enter the value that will act as an absolute threshold for the images, a value from 0 to 1.
Select the measurement to threshold with
(Used only if Measurement is selected for thresholding method)
Choose the image measurement that will act as an absolute threshold for the images.
Two-class or three-class thresholding?
(Used only for the Otsu thresholding method)
- Two classes: Select this option if the grayscale levels are readily
distinguishable into only two classes: foreground (i.e., regions of interest)
and background.
- Three classes: Choose this option if the grayscale
levels fall instead into three classes: foreground, background and a middle intensity
between the two. You will then be asked whether
the middle intensity class should be added to the foreground or background
class in order to generate the final two-class output.
Note that whether
two- or three-class thresholding is chosen, the image pixels are always
finally assigned two classes: foreground and background.
 Three-class thresholding may be useful for images in which you have nuclear staining along with
low-intensity non-specific cell staining. Where two-class thresholding
might incorrectly assign this intermediate staining to the nuclei
objects for some cells, three-class thresholding allows you to assign it to the
foreground or background as desired.
Three-class thresholding may be useful for images in which you have nuclear staining along with
low-intensity non-specific cell staining. Where two-class thresholding
might incorrectly assign this intermediate staining to the nuclei
objects for some cells, three-class thresholding allows you to assign it to the
foreground or background as desired.
 However, in extreme cases where either
there are almost no objects or the entire field of view is covered with
objects, three-class thresholding may perform worse than two-class.
However, in extreme cases where either
there are almost no objects or the entire field of view is covered with
objects, three-class thresholding may perform worse than two-class.
Assign pixels in the middle intensity class to the foreground or the background?
(Used only for three-class thresholding)
Choose whether you want the pixels with middle grayscale intensities to be assigned
to the foreground class or the background class.
Approximate fraction of image covered by objects?
(Used only when applying the MoG thresholding method)
Enter an estimate of how much of the image is covered with objects, which
is used to estimate the distribution of pixel intensities.
Method to calculate adaptive window size
(Used only if an adaptive thresholding method is used)
The adaptive method breaks the image into blocks, computing the threshold
for each block. There are two ways to compute the block size:
- Image size: The block size is one-tenth of the image dimensions,
or 50 × 50 pixels, whichever is bigger.
- Custom: The block size is specified by the user.
Size of adaptive window
(Used only if an adaptive thresholding method with a Custom window size
are selected)
Enter the window for the adaptive method. For example,
you may want to use a multiple of the largest expected object size.
Threshold correction factor
This setting allows you to adjust the threshold as calculated by the
above method. The value entered here adjusts the threshold either
upwards or downwards, by multiplying it by this value.
A value of 1 means no adjustment, 0 to 1 makes the threshold more
lenient and > 1 makes the threshold more stringent.
 When the threshold is calculated automatically, you may find that
the value is consistently too stringent or too lenient across all
images. This setting
is helpful for adjusting the threshold to a value that you empirically
determine is more suitable. For example, the
Otsu automatic thresholding inherently assumes that 50% of the image is
covered by objects. If a larger percentage of the image is covered, the
Otsu method will give a slightly biased threshold that may have to be
corrected using this setting.
When the threshold is calculated automatically, you may find that
the value is consistently too stringent or too lenient across all
images. This setting
is helpful for adjusting the threshold to a value that you empirically
determine is more suitable. For example, the
Otsu automatic thresholding inherently assumes that 50% of the image is
covered by objects. If a larger percentage of the image is covered, the
Otsu method will give a slightly biased threshold that may have to be
corrected using this setting.
Lower and upper bounds on threshold
Enter the minimum and maximum allowable threshold, a value from 0 to 1.
This is helpful as a safety precaution when the threshold is calculated
automatically, by overriding the automatic threshold.
 For example, if there are no objects in the field of view,
the automatic threshold might be calculated as unreasonably low; the algorithm will
still attempt to divide the foreground from background (even though there is no
foreground), and you may end up with spurious false positive foreground regions.
In such cases, you can estimate the background pixel intensity and set the lower
bound according to this empirically-determined value.
For example, if there are no objects in the field of view,
the automatic threshold might be calculated as unreasonably low; the algorithm will
still attempt to divide the foreground from background (even though there is no
foreground), and you may end up with spurious false positive foreground regions.
In such cases, you can estimate the background pixel intensity and set the lower
bound according to this empirically-determined value. - To view pixel intensities in an open image, use the
pixel intensity tool which is available in any open display window. When you move
your mouse over the image, the pixel intensities will appear in the bottom bar of the display window.
Select the smoothing method for thresholding
(Only used for strategies other than Automatic and
Binary image)
The input image can be optionally smoothed before being thresholded.
Smoothing can improve the uniformity of the resulting objects, by
removing holes and jagged edges caused by noise in the acquired image.
Smoothing is most likely
not appropriate if the input image is
binary, if it has already been smoothed or if it is an output of the
ClassifyPixels module.
The choices are:
- Automatic: Smooth the image with a Gaussian
with a sigma of one pixel before thresholding. This is suitable
for most analysis applications.
- Manual: Smooth the image with a Gaussian with
user-controlled scale.
- No smoothing: Do not apply any smoothing prior to
thresholding.
Threshold smoothing scale
(Only used if smoothing for threshold is Manual)
This setting controls the scale used to smooth the input image
before the threshold is applied. The scale should be approximately
the size of the artifacts to be eliminated by smoothing. A Gaussian
is used with a sigma adjusted so that 1/2 of the Gaussian's
distribution falls within the diameter given by the scale
(sigma = scale / 0.674)
Automatically calculate the size of objects for the Laplacian of Gaussian filter?
(Used only when applying the LoG thresholding method)
Select Yes to use the filtering diameter range above
when constructing the LoG filter.
Select No in order to manually specify the size.
You may want to specify a custom size if you want to filter
using loose criteria, but have objects that are generally of
similar sizes.
Enter LoG filter diameter
(Used only when applying the LoG thresholding method)
The size to use when calculating the LoG filter. The filter enhances
the local maxima of objects whose diameters are roughly the entered
number or smaller.
Method to distinguish clumped objects
This setting allows you to choose the method that is used to segment
objects, i.e., "declump" a large, merged object into individual objects of interest.
To decide between these methods, you can run Test mode to see the results of each.
-
Intensity: For objects that tend to have only a single peak of brightness
(e.g. objects that are brighter towards their interiors and
dimmer towards their edges), this option counts each intensity peak as a separate object.
The objects can
be any shape, so they need not be round and uniform in size as would be
required for the Shape option.
 This choice is more successful when
the objects have a smooth texture. By default, the image is automatically
blurred to attempt to achieve appropriate smoothness (see Smoothing filter options),
but overriding the default value can improve the outcome on
lumpy-textured objects.
This choice is more successful when
the objects have a smooth texture. By default, the image is automatically
blurred to attempt to achieve appropriate smoothness (see Smoothing filter options),
but overriding the default value can improve the outcome on
lumpy-textured objects. |
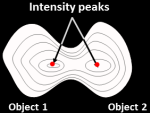 |
 The object centers are defined as local intensity maxima in the smoothed image.
The object centers are defined as local intensity maxima in the smoothed image.
-
Shape: For cases when there are definite indentations separating
objects. The image is converted to
black and white (binary) and the shape determines whether clumped
objects will be distinguished. The
declumping results of this method are affected by the thresholding
method you choose.
 This choice works best for objects that are round. In this case, the intensity
patterns in the original image are largely irrelevant. Therefore, the cells need not be brighter
towards the interior as is required for the Intensity option.
This choice works best for objects that are round. In this case, the intensity
patterns in the original image are largely irrelevant. Therefore, the cells need not be brighter
towards the interior as is required for the Intensity option. |
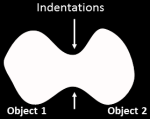 |
 The binary thresholded image is
distance-transformed and object centers are defined as peaks in this
image. A distance-transform gives each pixel a value equal to the distance
to the nearest pixel below a certain threshold, so it indicates the Shape
of the object.
The binary thresholded image is
distance-transformed and object centers are defined as peaks in this
image. A distance-transform gives each pixel a value equal to the distance
to the nearest pixel below a certain threshold, so it indicates the Shape
of the object.
- Laplacian of Gaussian: For objects that have an increasing intensity
gradient toward their center, this option performs a Laplacian of Gaussian (or Mexican hat)
transform on the image, which accentuates pixels that are local maxima of a desired size. It
thresholds the result and finds pixels that are both local maxima and above
threshold. These pixels are used as the seeds for objects in the watershed.
- None: If objects are well separated and bright relative to the
background, it may be unnecessary to attempt to separate clumped objects.
Using the very fast None option, a simple threshold will be used to identify
objects. This will override any declumping method chosen in the settings below.
Method to draw dividing lines between clumped objects
This setting allows you to choose the method that is used to draw the line
bewteen segmented objects, provided that you have chosen to declump the objects.
To decide between these methods, you can run Test mode to see the results of each.
Automatically calculate size of smoothing filter for declumping?
(Used only when distinguishing between clumped objects)
Select
Yes to automatically calculate the amount of smoothing
applied to the image to assist in declumping. Select
No to
manually enter the smoothing filter size.
This setting, along with the Minimum allowed distance between local maxima
setting, affects whether objects
close to each other are considered a single object or multiple objects.
It does not affect the dividing lines between an object and the
background.
Please note that this smoothing setting is applied after thresholding,
and is therefore distinct from the threshold smoothing method setting above,
which is applied before thresholding.
The size of the smoothing filter is automatically
calculated based on the Typical diameter of objects, in pixel units (Min,Max) setting above.
If you see too many objects merged that ought to be separate
or too many objects split up that
ought to be merged, you may want to override the automatically
calculated value.
Size of smoothing filter
(Used only when distinguishing between clumped objects)
If you see too many objects merged that ought to be separated
(under-segmentation), this value
should be lower. If you see too many
objects split up that ought to be merged (over-segmentation), the
value should be higher. Enter 0 to prevent any image smoothing in certain
cases; for example, for low resolution images with small objects
( < ~5 pixels in diameter).
Reducing the texture of objects by increasing the
smoothing increases the chance that each real, distinct object has only
one peak of intensity but also increases the chance that two distinct
objects will be recognized as only one object. Note that increasing the
size of the smoothing filter increases the processing time exponentially.
Automatically calculate minimum allowed distance between local maxima?
(Used only when distinguishing between clumped objects)
Select
Yes to automatically calculate the distance between
intensity maxima to assist in declumping. Select
No to
manually enter the permissible maxima distance.
This setting, along with the Size of smoothing filter setting,
affects whether objects close to each other are considered a single object
or multiple objects. It does not affect the dividing lines between an object and the
background. Local maxima that are closer together than the minimum
allowed distance will be suppressed (the local intensity histogram is smoothed to
remove the peaks within that distance). The distance can be automatically
calculated based on the minimum entered for the
Typical diameter of objects, in pixel units (Min,Max) setting above,
but if you see too many objects merged that ought to be separate, or
too many objects split up that ought to be merged, you may want to override the
automatically calculated value.
Suppress local maxima that are closer than this minimum allowed distance
(Used only when distinguishing between clumped objects)
Enter a positive integer, in pixel units. If you see too many objects
merged that ought to be separated (under-segmentation), the value
should be lower. If you see too many objects split up that ought to
be merged (over-segmentation), the value should be higher.
The maxima suppression distance
should be set to be roughly equivalent to the minimum radius of a real
object of interest. Any distinct "objects" which are found but
are within two times this distance from each other will be assumed to be
actually two lumpy parts of the same object, and they will be merged.
Speed up by using lower-resolution image to find local maxima?
(Used only when distinguishing between clumped objects)
Select
Yes to down-sample the image for declumping. This can be
helpful for saving processing time on large images.
Note that if you have entered a minimum object diameter of 10 or less, checking
this box will have no effect.
Retain outlines of the identified objects?
Select Yes to retain the outlines of the new objects
for later use in the pipeline. For example, a common use is for quality control purposes by
overlaying them on your image of choice using the OverlayOutlines module and then saving
the overlay image with the SaveImages module.
Name the outline image
(Used only if the outline image is to be retained for later use in the pipeline)
Enter a name for the outlines of the identified
objects. The outlined image can be selected in downstream modules by selecting
them from any drop-down image list.
Fill holes in identified objects?
Select
After both thresholding and declumping to fill in background holes
that are smaller than the maximum object size prior to declumping
and to fill in any holes after declumping.
Select
After declumping only to fill in background holes
located within identified objects after declumping.
Select Never to leave holes within objects.
Please note that if a foreground object is located within a hole
and this option is enabled, the object will be lost when the hole
is filled in.
Handling of objects if excessive number of objects identified
This setting deals with images that are segmented
into an unreasonable number of objects. This might happen if
the module calculates a low threshold or if the image has
unusual artifacts.
IdentifyPrimaryObjects can handle
this condition in one of three ways:
- Continue: Don't check for large numbers
of objects.
- Truncate: Limit the number of objects.
Arbitrarily erase objects to limit the number to the maximum
allowed.
- Erase: Erase all objects if the number of
objects exceeds the maximum. This results in an image with
no primary objects. This option is a good choice if a large
number of objects indicates that the image should not be
processed.
Maximum number of objects
(Used only when handling images with large numbers of objects by truncating)
This setting limits the number of objects in the
image. See the documentation for the previous setting
for details.
 Our recommendation or example use case for which a particular setting is best used.
Our recommendation or example use case for which a particular setting is best used. Indicates a condition under which a particular setting may not work well.
Indicates a condition under which a particular setting may not work well. Technical note. Provides more detailed information on the setting.
Technical note. Provides more detailed information on the setting.
