Once the images have been identified with the Images module, have had metadata associated with them using the Metadata module, and have been assigned names by the NamesAndTypes module, you have the option of further sub-dividing the image sets into groups that share a common feature. Some downstream modules of CellProfiler are capable of processing groups of images in useful ways (e.g., object tracking within a set of images comprising a time series, illumination correction within a set of images comprising an experimental batch, data export for a set of images comprising a plate).
What is an image "group"?
The key to understanding why grouping may be necessary is that CellProfiler processes the input images sequentially and in the order given by the NamesAndTypes module. If you have multiple collections (or "groups") of images that should be processed independently from each other, CellProfiler will simply finish processing one collection and proceed to the next, ignoring any distinction between them unless told otherwise via the Groups module.
To illustrate this idea, below are two examples where the grouping concept can be useful or important:
- If you have time-lapse movie data that is in the form of individual image files, and you are performing object tracking, it is important to indicate to CellProfiler that the end of a movie indicates the end of a distinct data set. Without doing so, CellProfiler will simply take the first frame of the next movie as a continuation of the previous one. If each set of files that comprise a movie is defined using the Metadata module, the relevant metadata can be used in this module to insure that object tracking only takes place within each movie.
- If you are performing illumination correction for a screening experiment, we recommend that the illumination function (an image which represents the overall background fluorescence) be calculated on a per-plate basis. Since the illumination function is an aggregate of images from a plate, running a pipeline must yield a single illumination function for each plate. Running this pipeline multiple times, once for each plate, will give the desired result but would be tedious and time-consuming. In this case, CellProfiler can use image grouping for this purpose; if plate metadata can be defined by the Metadata module, grouping will enable you to process images that have the same plate metadata together.
What are the inputs?
Using this module assumes that you have already adjusted the following Input modules:- Used the Images module to produce a list of images to analyze.
- Used the Metadata module to produce metadata defining the distinct sub-divisions between groups of images.
- Used the NamesAndTypes module to assign names to individual channels and create image sets.
What do the settings mean?
See below for help on the individual settings. Selecting this module will display a panel, allowing you to select whether you want to create groups or not. A grouping may be defined as according to any or as many of the metadata categories as defined by the Metadata module. By selecting a metadata tag from the drop-down for the metadata category, the Groups module will sub-divide and assemble the image sets according to their unique metadata value. Upon adding a metadata category, the two tables underneath will update to show the resultant organization of the image sets for each group.What do I get as output?
The final product of the Groups module is a list defining subsets of image sets that will be processed independently of the other subsets.- If no groups are defined, the Analysis modules in the rest of the pipeline will be applied to all images in exactly the same way.
- If groups are defined in the Groups module, then organizationally (and transparently to you), CellProfiler will begin the analyses with the first image set of the group, end with the last image set of the group, and then proceed to the next group.
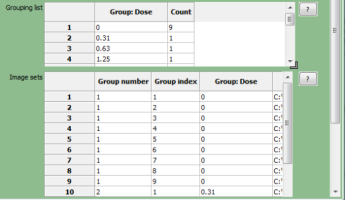
The two tables at the bottom provide the following information when a metadata category is selected:
- The grouping list(top table) shows the unique values of the selected metadata under the "Group" column; each of the unique values comprises a group. The "Count" column shows the number of image sets included in a given group; this is useful as a "sanity check" to make sure that the expected numbers of images are present.
- The image set list (bottom table) shows the file name and location of each of the image sets that comprise the groups.
 |
Available measurements
- Group_Number: The index of each grouping, as defined by the unique combinations of the metadata tags specified. These are written to the per-image table.
- Group_Index: The index of each imaget set within each grouping, as defined by the Group_Number. These are written to the per-image table.